SICB Annual Meeting 2020
January 3-7, 2020
Austin, TX
Symposium S7: Building Bridges from Genome to Phenome: Molecules, Methods and Models
Understanding the mechanistic basis by which genes give rise to both stability and variation in phenotypes is one of the Grand Challenges articulated by NSF. Accordingly, scientists in many disciplines are using a wide variety of organisms and experimental approaches to generate complex datasets at different levels of biological organization, all aiming to elucidate aspects of the genome-to-phenome framework. The goals of Building Bridges Symposium and related oral and poster complementary sessions are to build connections among the most successful of these research efforts, as well as to identify critical gaps and future needs to accelerate and facilitate this critical area of research. The Symposium is organized by members of NSF’s Animal Genome-to-Phenome Research Coordination Network (AG2P RCN) to encourage networking across disciplines, gender, ethnicity, and professional ranks. Participant discussions at a concluding workshop will serve as the basis for a white paper identifying major gaps, key barriers and leading edges in the field of genome-to-phenome research.
Sponsors: SICB Co-Sponsoring Divisions DCE, DCPB, DEDB, DEDE, DEE, DIZ, DPCB
AMS, TCS
AG2P RCN

Organizers
- Karen G. Burnett, Research Affiliate, Department of Biology, College of Charleston,
- David S. Durica, Professor Emeritus, Department of Biology, University of Oklahoma,
- Donald L. Mykles, Professor, Department of Biology, Colorado State University
- Jonathon Stillman, Professor, Department of Biology, San Francisco State University
Speakers
S7-1 Monday, Jan. 6, 07:45 BURNETT, KG*; DURICA, DS; MYKLES, DL; STILLMAN, JH:
SICB Wide Symposium: Building Bridges from Genome to Phenome: Molecules, Methods and Models
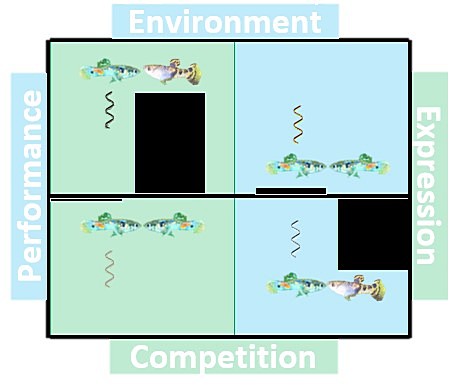
S7-2 Monday, Jan. 6, 08:00 MAURO, AA*; GHALAMBOR, CK; MAURO, Alexander:
The transcriptomic basis of a trade-off between salinity tolerance and competitive ability in the Trinidadian guppy
S7-3 Monday, Jan. 6, 08:30 MYKLES, D:
Applying transcriptomics and proteomics to study phenotypic transitions of the crustacean molting gland (Y-organ)
S7-4 Monday, Jan. 6, 09:00 LI, J*; LEVITAN, B B; KÃœLTZ, D:
Quantitation and comprehension of context-dependent changes of dynamic proteomes
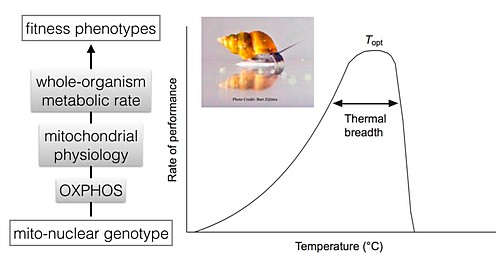
S7-5 Monday, Jan. 6, 10:00 HAVIRD, JC:
Building Bridges from the Mitogenome to the Mitophenome to the Organismal Phenome
S7-6 Monday, Jan. 6, 10:30 SHARBROUGH, J*; MONTOOTH, K; NEIMAN, M:
Phenotypic Variation in Mitochondrial Function across New Zealand Snail Populations
S7-7 Monday, Jan. 6, 11:00 SANTOS, SR*; HOFFMAN, SK; SEITZ, KW; HAVIRD, JC; WEESE, DA:
Phenotypic Comparability Arising from Genotypic Variability amongst Physically Structured Microbial Consortia
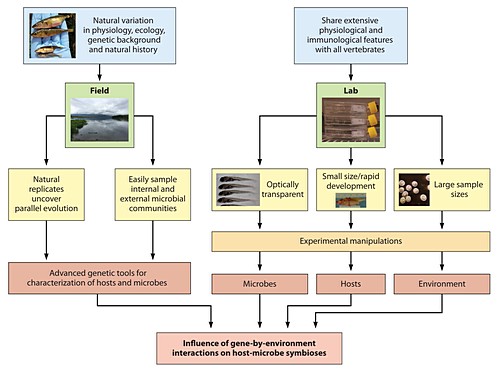
S7-8 Monday, Jan. 6, 11:30 MILLIGAN-MYHRE, KCA:
Using an evolutionary model organism to reveal host genetic influence on host-microbe interactions
S7-9 Monday, Jan. 6, 13:30 SCHAEFER, RJ*; BAXTER, I; MCCUE, ME:
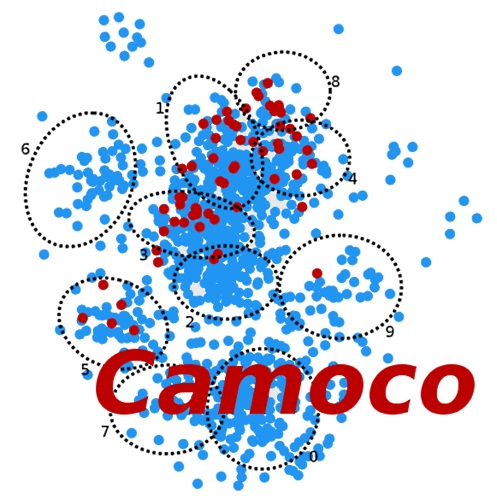
Using Camoco to integrate genome-wide association studies with context specific co-expression networks in corn and horses
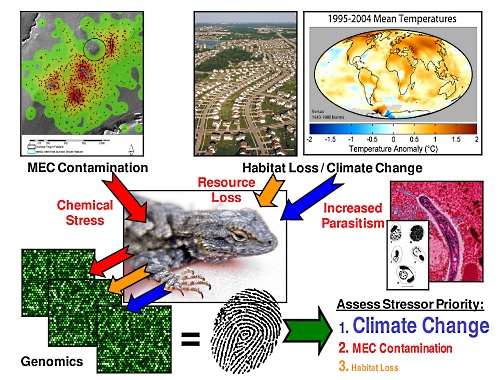
S7-10 Monday, Jan. 6, 14:00 GUST, KA:
Omics in Non-Model Species: Closing the Loop Among Genes, Molecular Systems, and Phenotypes to predict Adverse Outcomes to Environmental Stress
S7-11 Monday, Jan. 6, 14:30 HAHN, DA; HAHN, Daniel:
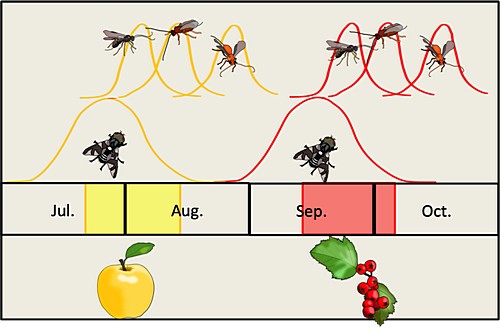
Combining ‘omics Approaches to Pick Apart the Genetic and Physiological Architectures of Seasonal Adaptation
S7-12 Monday, Jan. 6, 15:00 GARRETT, AD; BRENNAN, RS; STEINHART, A; PELLETIER, A; PESPENI, MH*:
Linking Genome to Phenome for Complex Traits: Studies of Global Change Adaptive Variation in Marine Invertebrates